| Location: Home > Papers |
| First Author: | ZHENG Fei |
| Abstract: |
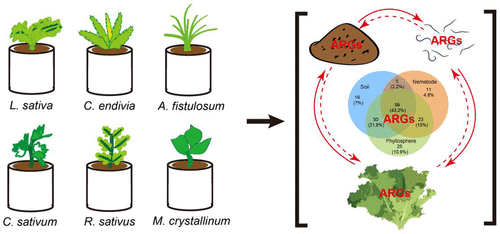
Resistomes are ubiquitous in natural environments. Previous studies have shown that both the plant phyllosphere and soil-borne nematodes were reservoirs of above- and below-ground resistomes, respectively. However, the influence of plant identity on soil, nematode, and phyllosphere resistomes remains unclear. Here, a microcosm experiment was used to explore the characteristics of bacterial communities and resistomes in soil, nematode, and phyllosphere associated with six different plant identities (Lactuca sativa, Cichorium endivia, Allium fistulosum, Coriandrum sativum, Raphanus sativus, and Mesembryanthemum crystallinum). A total of 222 antibiotic resistance genes (ARGs) and 7 mobile genetic elements (MGEs) were detected by high-throughput quantitative PCR from all samples. Plant identity not only significantly affected the diversity of resistomes in soil, nematode, and phyllosphere but also influenced the abundance of resistomes in nematodes. Shared bacteria and resistomes indicated a possible pathway of resistomes transfer through the soil–nematode–phyllosphere system. Structural equation models revealed that plant identity had no direct effect on phyllosphere ARGs, but altered indirectly through complex above- and below-ground interactions (soil-plant-nematode trophic transfer). Results also showed that bacteria and MGEs were key factors driving the above- and below-ground flow of resistomes. The study extends our knowledge about the top-down and bottom-up dispersal patterns of resistomes.
|
| Contact the author: | YANG Xiao-Ru |
| Page Number: | |
| Issue: | |
| Subject: | |
| Impact Factor: | |
| Authors units: | |
| PubYear: | August 2, 2022 |
| Volume: | |
| Publication Name: | Environmental Science & Technology |
| The full text link: | https://doi.org/10.1021/acs.est.1c08733 |
| ISSN: | |
| Appendix: |