Microeukaryotic plankton constitute part of the base of aquatic food webs and play important roles in the structure and function of aquatic ecosystems. Reservoir microeukaryotic plankton (0.2–200 microns) including protozoa, algae, fungi and small metazoan, etc., are characterized by their small size and high diversity. This diversity has been studied through light microscopy since the work of the early microscopists of the European Enlightenment. However, there are clear limitations in this approach. Some extremely small eukaryotes are difficult to observe or distinguish by light microscopy, and in addition, morphologically similar individuals may belong to different cryptic species or alternatively some morphologically distinct types may actually be from the same species. To address this it is now common to use SSU rRNA gene-based approaches which can successfully described and characterized microeukaryotic plankton in aquatic systems, but it is difficult to distinguish the size of microeukaryotic plankton. Previous molecular studies of microeukaryotic diversity have often used size-fractionated samples (where microbes of different sizes are separated by sieving) to separate microeukaryotic plankton from large organisms, an approach which has long been used with great success in traditional microscopy-based plankton studies. However, we still lack sufficient understanding about whether traditional size-fraction filtering can accurately and effectively discriminate the microeukaryotic plankton communities from large eukaryotic communities in gene-based studies.
Researchers in Aquatic Ecohealth Group (Jun Yang’s research team), Institute of Urban Environment, Chinese Academy of Sciences, in collaboration with Dave Wilkinson at Lincoln University UK, studied the eukaryotic plankton communities from two subtropical reservoirs in Xiamen city, southeast China based on high-throughput sequencing of 18S rDNA amplicons and size-fraction filtering approach. They found that the filtering had little effect on the results with species richness, alpha and beta diversities of plankton communities and relative abundance of 99.2% eukaryotic OTUs showing no significant changes after prefiltering by a 200 μm pore-size sieve in the two subtropical reservoirs. Furthermore, about 25% reads were classified as metazoa in both size-fraction groups. They further found that both >0.2 μm and 0.2–200 μm eukaryotic plankton communities exhibited very similar, and synchronous, spatiotemporal patterns and processes associated with almost identical environmental drivers. This was most likely due to 200 μm pore-size filtering, which is unable to effectively remove irregular body forms of large eukaryotes and fragments of DNA in the water (environmental DNA, eDNA). In addition, their results also highlight the importance of sequencing depth, and strict quality filtering of reads, when designing studies to characterize microeukaryotic plankton communities. They concluded that SSU rDNA-based size-fractionated filtering approach failed to avoid eDNA contamination. Therefore, more effective and complementary approaches should be considered as a supplement for future ecology studies on microeukaryotic plankton such as microscopy and SSU rRNA sequencing rather than just relying on rDNA-based molecular approaches. These findings were recently published in Molecular Ecology Resources (Lemian Liu, Min Liu, David M Wilkinson, Huihuang Chen, Xiaoqing Yu and Jun Yang*. 2017. DNA metabarcoding reveals that 200-μm-size-fractionated filtering is unable to discriminate between planktonic microbial and large eukaryotes, doi: 10.1111/1755-0998.12652, http://onlinelibrary.wiley.com/doi/10.1111/1755-0998.12652/full).
This work was funded by the National Natural Science Foundation of China, the Natural Science Foundation for Distinguished Young Scholars of Fujian Province and the Knowledge Innovation Program of the Chinese Academy of Sciences.
Key words: eDNA, eukaryotic planktonic community, high-throughput sequencing, size-fractionated filtering, subtropical reservoir
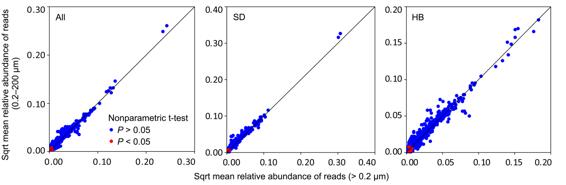
Figure captions: Almost all (>99%) OTUs showed no significant difference in relative abundance of reads between the >0.2 μm and 0.2 to 200 μm eukaryotic plankton communities in both reservoirs (All), in Shidou Reservoir (SD) and in Hubian Reservoir (HB).
Corresponding author: Professor Jun YANG,
Institute of Urban Environment, Chinese Academy of Sciences
http://www.iue.ac.cn/ 1799
Jimei Road, Xiamen 361021, China
Tel/Fax: 86 592 6190775;
Email: jyang@iue.ac.cn