Scientists investigated the similar but not identical resuscitation trajectories of the soil microbial community based on either DNA or RNA after flooding
Scientists from the Institute of Urban Environment, Chinese Academy of Sciences, have detected and discussed the similar but not identical resuscitation trajectories of the soil microbial community based on either DNA or RNA after flooding.
Soil moisture is a crucial determinant of soil microbial activities and ecosystem functions. Not only drought but also flooding are unfavorable for soil microorganisms to metabolize and maximize the production and of energy. The switch of soil water status from drought to flooding can happen rapidly and microbial activity might be either stimulated or further inhibited, but we still have insufficient understanding of the underlying microbial processes.
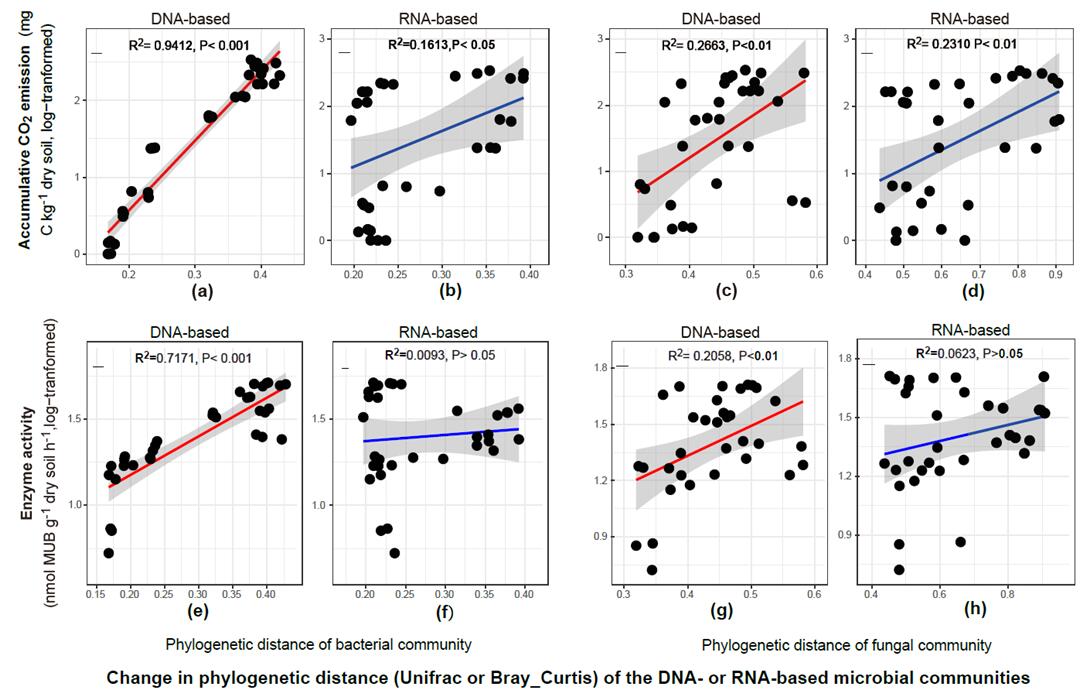
Here, we tracked the changes in soil bacterial and fungal abundance and their community structures based on high-throughput sequencing from amplicons of rRNA genes or transcripts, assaying the total (DNA-based) and potentially active (RNA-based) communities in response to abrupt flooding of dry soil. Also, the changing trajectories of the soil microbial community were analyzed after flooding. Results showed that the bacterial community was found to be more responsive than the fungal community to flooding, and the accumulative amount of CO2 released was more closely related than enzyme activity to the change in structure of the bacterial community after flooding. According to the clustering of bacterial species over the time series, the bacterial community responses were clearly classified into three distinct patterns (rapid, intermediate and delayed) in which the intermediate responders dominated by Bacilli displayed highly phylogenetic clustering, suggesting the potential importance of Bacilli in the microbial assemblage and subsequent ecosystem functioning.
The study was published in Agronomy entitled " Similar but Not Identical Resuscitation Trajectories of the Soil Microbial Community Based on Either DNA or RNA after Flooding".
This study was supported by the National Natural Science Foundation of China.
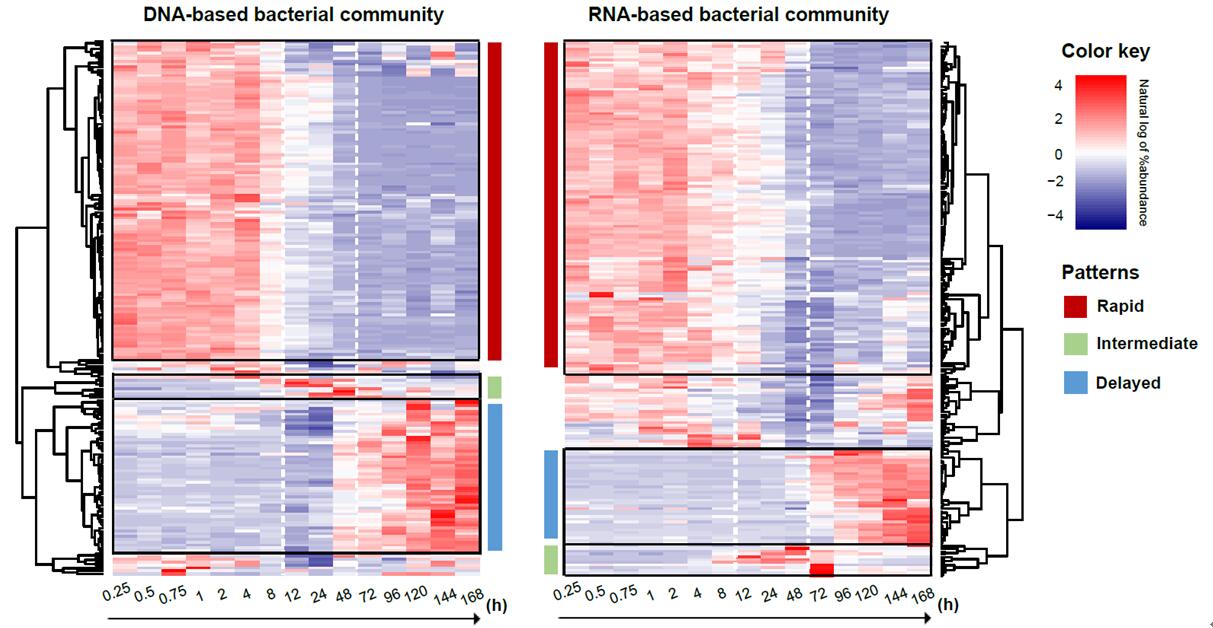
Heatmaps displaying the changes in the relative abundance of bacterial genera with abundance > 0.1% based on (a) DNA and (b) RNA community analysis over the time series. A similar trend in changes in the relative abundance and clustering of bacterial taxa were found over time. In both DNA- and RNA-based analysis, bacterial genera are significantly classified into three different responding patterns (dark red: rapid pattern; light green: intermediate pattern; light blue: delayed pattern) according to the changes of relative abundance over the time series.