Since antibiotics are commonly used in livestock industry in developing countries, animal waste has become an important reservoir of antibiotic resistance genes (ARGs). ARGs receive more public concern because they can transfer among different hosts by horizontal gene transfer (HGT) and pose a risk to human health. Evidences suggest that long-term application of manure and manure-derived fertilizer can increase antibiotic resistance in agricultural soils. These findings highlight the need to critically evaluate the potential of manure treatment processes to attenuate ARGs and limit their spread to the environment.
Prior to land application, biotechnologies such as aerobic composting and anaerobic digestion have been broadly employed to treat animal waste. Of them, anaerobic digestion can not only degrade organic matter and remove pathogen effectively, but also produce renewable energy economically. Recent studies have begun to investigate the fate of ARGs during anaerobic digestion of manure. Most of these studies investigated a handful of ARGs. Critical factor for the removal of ARGs and underlying mechanisms have not reached consistent conclusions.
Supported by the National Natural Science Foundation of China and the Knowledge Innovation Program of the Chinese Academy of Sciences, the Liu chaoxiang team in Institute of Urban Environment, Chinese Academy of Sciences employed high throughput quantitative PCR (HT-qPCR) targeting almost all major classes of ARGs and mobile genetic elements (MGEs) to characterize the dynamics of ARGs profile in anaerobic digestion of swine manure. The effects of temperature and residual antibiotics on ARGs variation were emphatically analyzed. Meanwhile, the shift of microbial communities was comprehensively explored using illumina sequencing of 16S rRNA gene. The contribution of experimental conditions, chemical properties and microbial taxa to the variation of ARGs and the co-occurrence patterns between ARGs and microbial community were analyzed thereafter.
This research has been entitled as “Higher Temperatures Do Not Always Achieve Better Antibiotic Resistance Gene Removal in Anaerobic Digestion of Swine Manure” and published in Applied and Environmental Microbiology (AEM). Associate professor Xu Huang is the first author and corresponding author. Professor Chaoxiang Liu is the co-corresponding author.
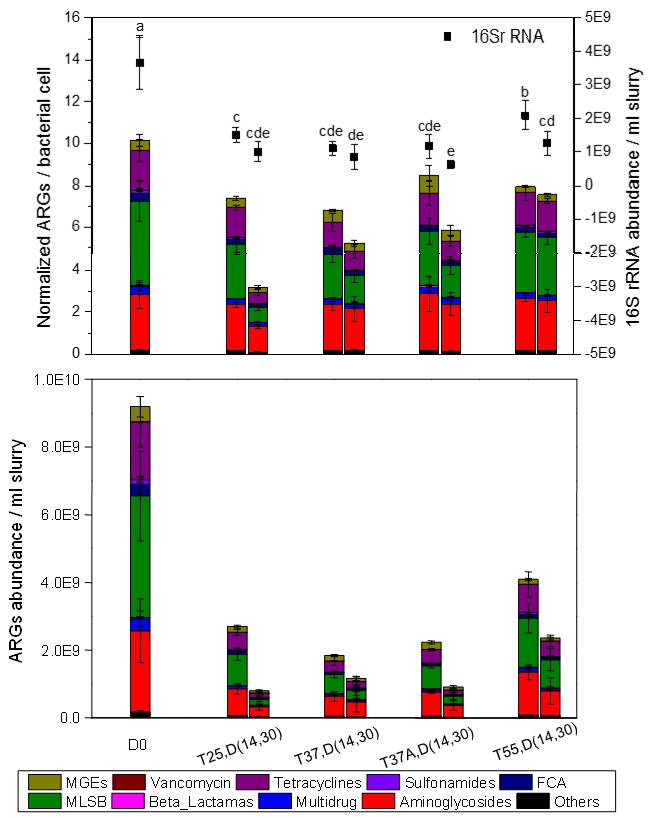
Fig. 1 Abundance of ARGs, MGEs and 16S rRNA gene detected in manure slurry on days 0, 14 and 30. In sample name, the letters “D”, “T” and “A” indicate day, temperature and antibiotics respectively.
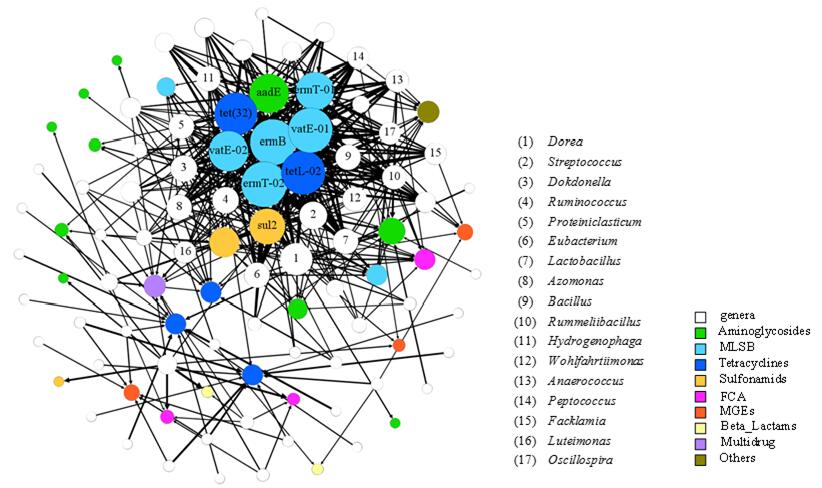
Fig. 2 Network analysis showing the significant correlation between 64 microbial genera (source) and 35 ARGs (target) (p < 0.01).